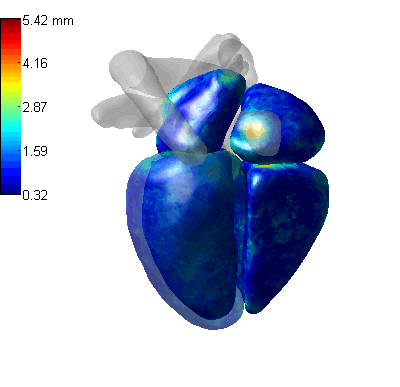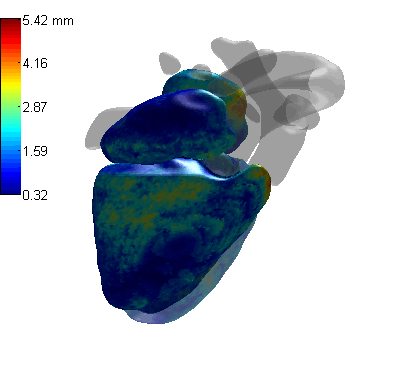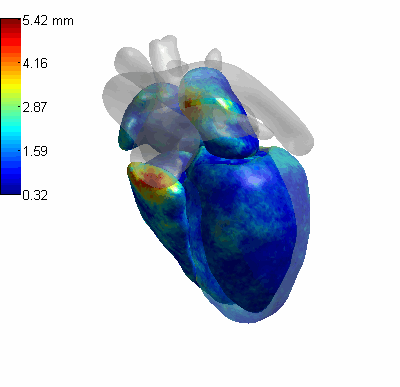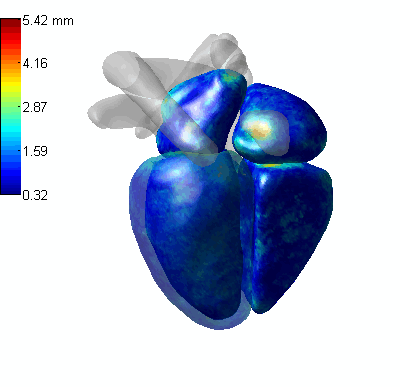
3D CoordConv Segmentation
Grand Challenge 2017 Multi-Modality Whole Heart Segmentation
- http://www.sdspeople.fudan.edu.cn/zhuangxiahai/0/mmwhs/
Contribution
- An Intriguing Failing of Convolutional Neural Networks and the CoordConv Solution [https://arxiv.org/abs/1807.03247]
Training Run
In code directory
> python main.py --params=ct_train.json
Result
| Model | Background | MLV | LABC | LVBC | RABC | RVBC | ASA | PUA | Average DSC |
|---|---|---|---|---|---|---|---|---|---|
| U-net 3D | 0.995 | 0.918 | 0.929 | 0.912 | 0.925 | 0.923 | 0.843 | 0.923 | 0.909 |
| U-net 3D + CoordConv | 0.995 | 0.919 | 0.926 | 0.912 | 0.933 | 0.924 | 0.928 | 0.897 | 0.920 |
- MLV: the Myocardium of the left ventricle, LABC: the left atrium blood cavity, LVBC: the left ventricle blood cavity, RABC: the right atrium blood cavity, RVBC: the right ventricle blood cavity, ASA: the ascending aorta, PUA: the pulmonary artery
- Average DSC is average of classes that excluded background
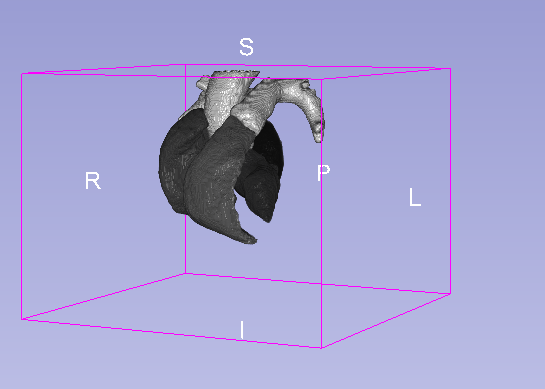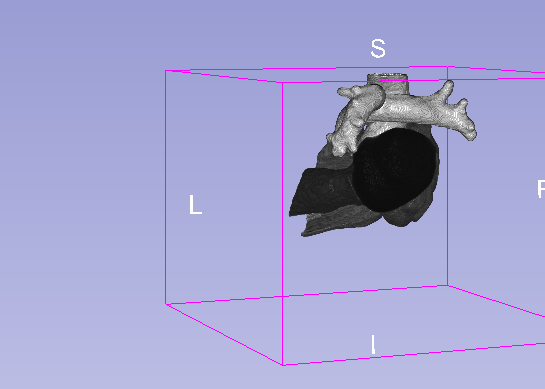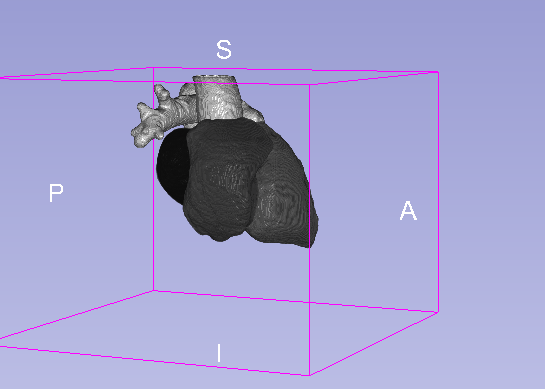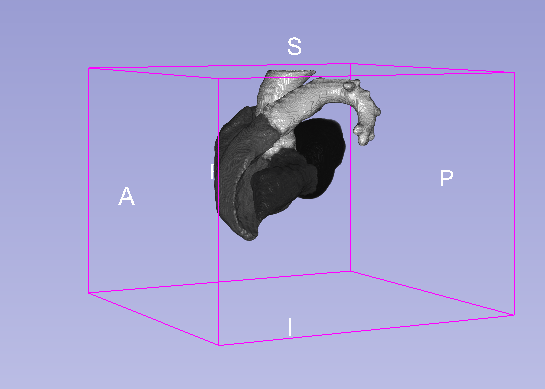
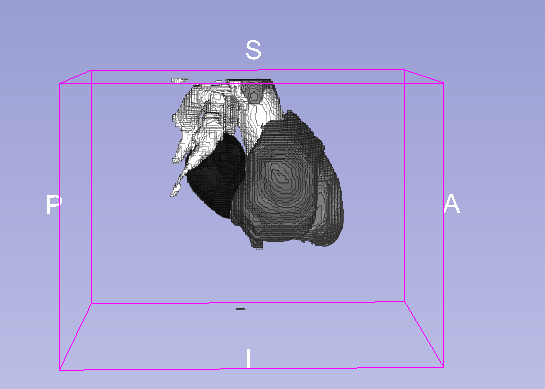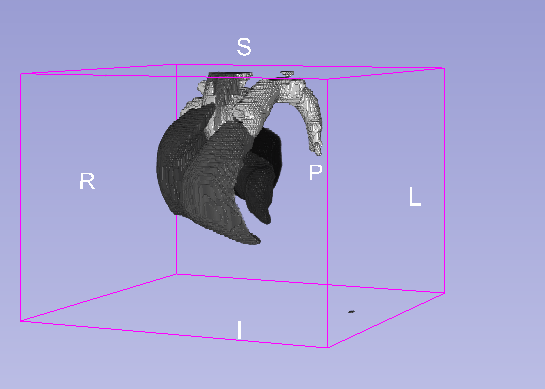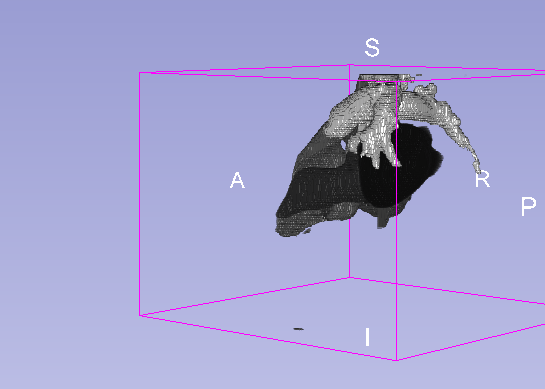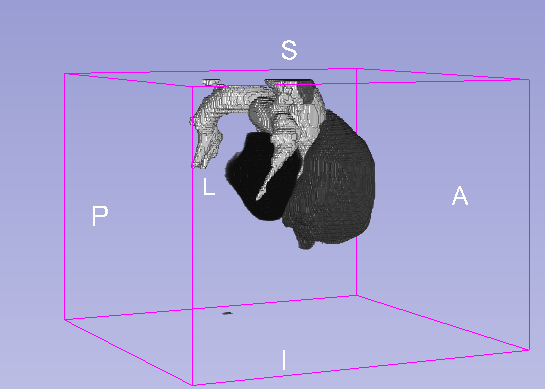
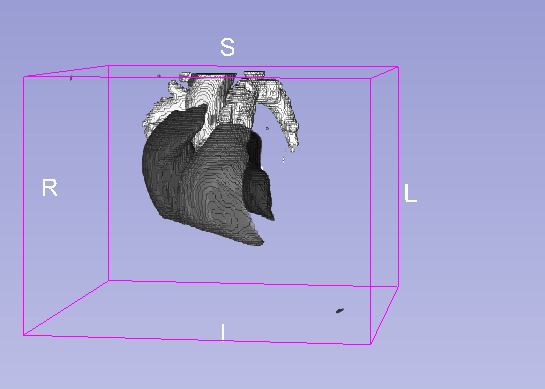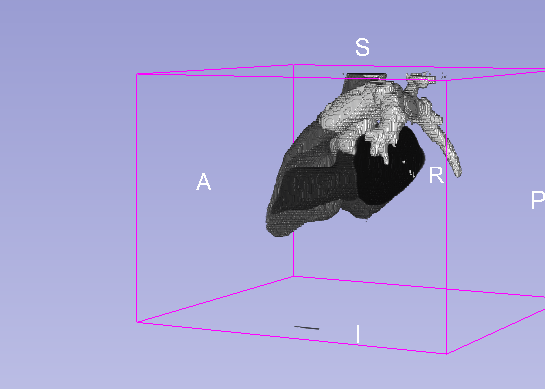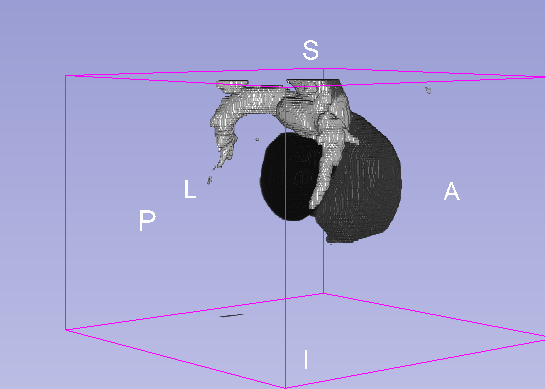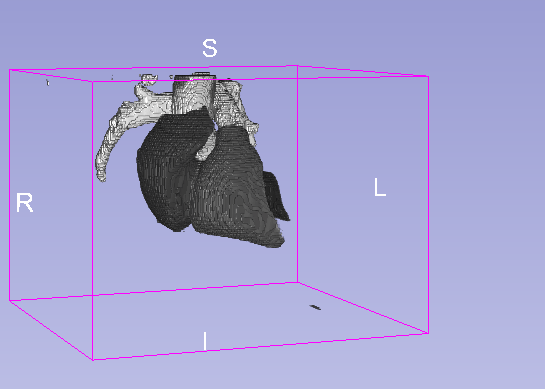
Details
| Data | Number of train set | Number of validation set | Patch dim | Resize rate | Batch size | Epochs | Number of train patch image | Number of validation patch image | Metric | Loss function | Optimizer | Learning rate | Number of GPU |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CT | 18 | 2 | 96 | 0.7 | 2 | 100 | 20 | 100 | Dice Similarity Coefficient | dice coefficient loss | Adam | 0.0001 | 4 |
Limit
The host server is down, so the test set can no longer be evaluated.